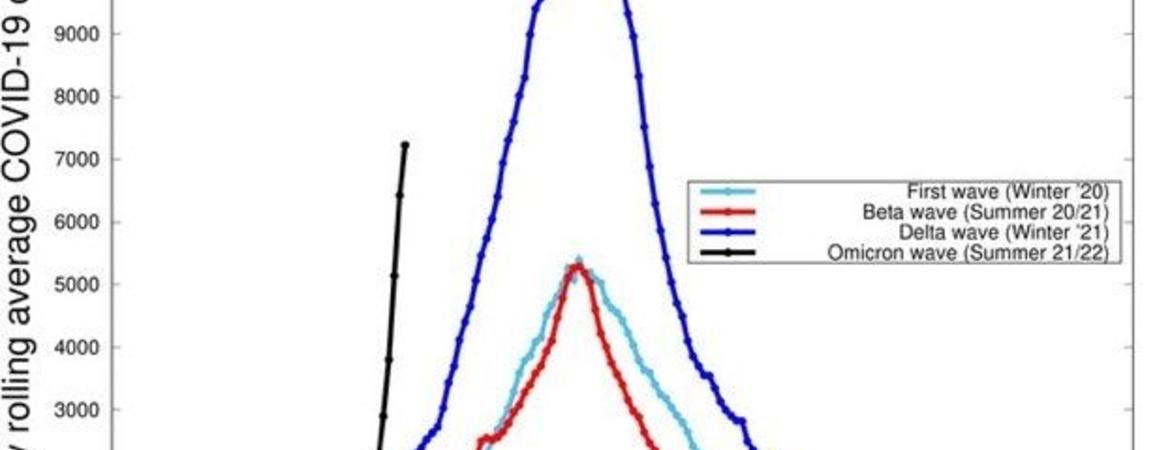
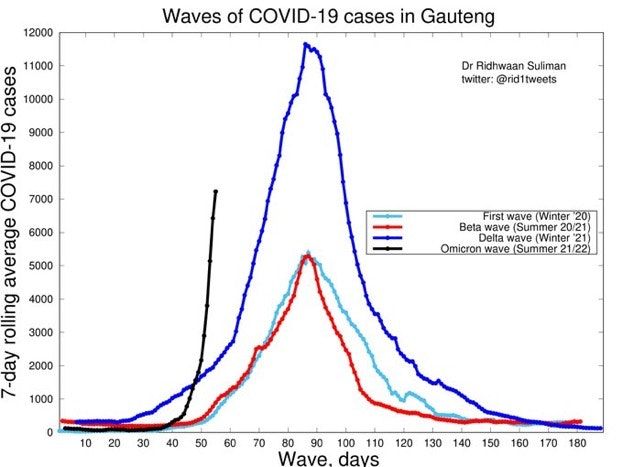
Omicron, aka B.1.1.529, is the latest Covid Variant of Concern (VoC) and carries far more mutations than earlier variants, including 32 mutations in the spike protein gene alone. And this news of large numbers of mutations, combined with the scary forecast of Omicron’s dramatically increased infectivity (see case count graph from South Africa, below) and putative antibody avoidance, is definitely worrisome, if not panic inducing.
But what’s the significance of the quantity of mutations? Fortunately, the answer is, not much. The number of mutations is immaterial; the effect of each mutation, singly or in combination, is what matters. Unfortunately, several of the mutations do make Omicron more scary.
What is a mutation? A mutation is defined simply as a permanent (heritable) change in the DNA (or, in the case of Coronavirus, RNA) base sequence. A mutation could be a substitution of one base for another (e.g. Adenine (A) to Guanine (G)), a deletion of one or more bases, an insertion of one or more bases, a duplication of one of more bases, an inversion of a block of bases, or just about any other base rearrangement you might imagine. A mutation is manifest when the base sequence in a given gene also changes the amino acid sequence of the corresponding protein. A gene can be thought of as a recipe for a protein, and the presence of that specific protein provides the trait or feature. When the base sequence of the gene changes, that can result in a change in the corresponding amino acid in the protein. The cell machinery responsible for protein synthesis reads the DNA (or RNA) base sequence three letters at a time (called a triplet codon) and constructs a growing chain of amino acids as directed from the three base letter instruction or “code”. There are 20 different amino acids in proteins, so substituting one amino acid for another can (but does not always) result in a change of some feature of the protein.
For example, the DNA ( or RNA) base sequence GAG calls for the amino acid Glutamic Acid, which the cell machinery places into the growing chain. If a mutation changes the first G to A, the resulting triplet sequence, AAG, results in the amino acid Lysine being placed into the protein where the Glutamic Acid was. Depending on the exact location of this amino acid substitution, the resulting protein may be unaffected, or have some feature altered, or even rendered completely non-functional.
Here’s a crucial and often misunderstood fact: Mutations are not directed, but random. That is, the virus has no means of predicting and directing a particular mutation that might improve its infectivity. A mutation occurs randomly and spontaneously in one virus particle in a population of viruses, then life goes on, with the mutation either improving infectivity, reducing infectivity, or having no effect on infectivity of that one virus particle in the surrounding population of virus particles. If the random, spontaneous mutation does improve infectivity, that mutated virus particle enjoys an adaptive advantage over all the other nearby virus that didn’t mutate. As a result of this infectious improvement, the progeny of the “improved” virus will, over time, come to dominate the population of viruses. At this point, scientists might notice the genetically different viral strain and give it a name to distinguish it from the initial strain.
While a large number of mutations may increase the likelihood of carrying at least one which increases pathogenicity, the same probability increases the likelihood of diminished pathogenicity. That is, one specific mutation might result in a spike protein better able to bind to the human host cell ACE-2 protein to facilitate infection, thus increasing pathogenicity. But another specific mutation, perhaps simultaneously, might reduce the spike protein’s affinity for the host cell’s ACE-2 protein, thus reducing pathogenicity. With Omicrons 32 mutations on the spike gene, some could well increase infectivity while others could inhibit infectivity. Most mutations have no discernable clinical impact, however.
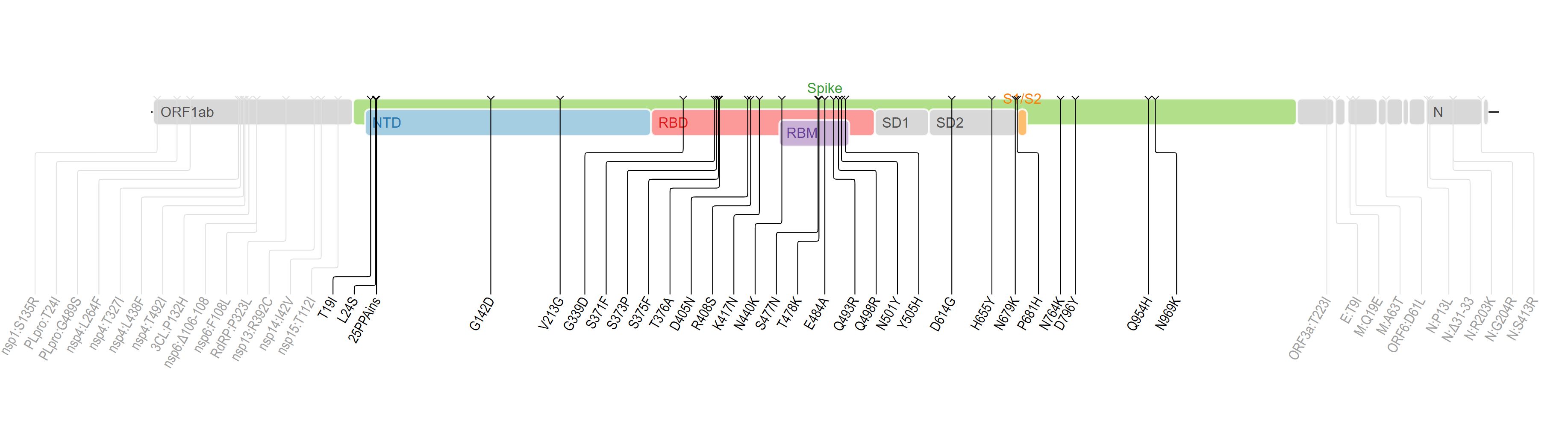
Omicron's mutations are shown in the map image below. The Spike gene, in green, has a relatively large number of mutations that result in changes to the amino acid sequence of the final spike protein. For example, the 440th amino acid in the spike protein is mutation N440K, in which the original amino acid Asparagine (abbreviated N) is replaced with Lysine (K). This single amino acid change, also observed in other variant strains, is thought to substantially increase infectivity. A little further along the amino acid chain, at position 484, is mutation E484K (also known as the Eek" mutation). Here, the amino acid Glutamic acid (abbreviated 'E') is replaced by the amino acid Lysine ('K') as the 484th amino acid in the spike protein chain. This particular mutation, also present in some other variant stains, is thought to increase the virus's ability to evade antibodies. Although these mutations, and others, are also present in other SARS-CoV-2 variant strains, the combination of different mutations results in Omicron gaining multiple tools to increase its virulence, ranging from increased infectivity to enhanced ability to evade antibodies or other protective measures the body uses to fight off pathogens.
To initiate an infection, the SARS-CoV-2 virus spike protein is attracted to ACE-2, a protein on the surface of the target host cell. If the ACE-2 protein and the spike protein fit together like a key in a lock, that opens a door to the host cell and the infection proceeds. Once inside the cell, the viral genome hijacks the cell machinery to make more copies of the virus, which eventually distribute to infect other cells and ultimately, other human targets. Mutations in the spike gene can result in a more effective "key" for the ACE-2 "lock", resulting in increased infectivity. This is why it’s so important to limit viral infections as soon as possible, because multiplying the virus leads to both more mutations and more human victims. The most effective way to limit viral infections is through vaccinations, masking in public and social distancing.
Citations and further reading
Mullen J, Tsueng G, et int., and tCfVSB. “Outbreak.info.” outbreak.info, 2020. - https://covdb.stanford.edu/page/mutation-viewer/#omicronOutbreak.info B.1.1.529 Lineage Reportb-1-1-529_1638019891935.svg, CC BY-SA 4.0,
https://commons.wikimedia.org/w/index.php?curid=112859849
https://covdb.stanford.edu/page/mutation-viewer/
https://en.wikipedia.org/wiki/Variants_of_SARS-CoV-2
https://en.wikipedia.org/wiki/SARS-CoV-2_Omicron_variant
https://yourlocalepidemiologist.substack.com/p/omicron-were-getting-some-answers
https://www.nytimes.com/interactive/2021/health/coronavirus-variant-tracker.html